Analysis Protein Drugs Design Program with Quality and Structure Prediction
DOI:
https://doi.org/10.31033/abjar.2.2.1Keywords:
development, design, proteinAbstract
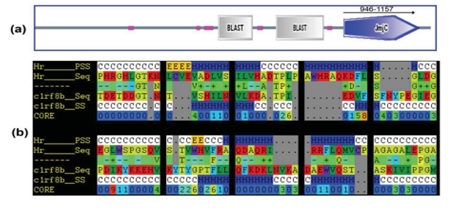
A complete lack of hair will result when the "hairless protein" associated with the hairless gene, which is required for hair growth, ceases to function. The coordinates of this gene are 22027873-22045326 on chromosome 8. The JmjC domain superfamily includes the hairless gene, which also contributes to the process of histone demethylation. The domain sequence is 212 amino acids long and spans residue positions 946 to 1157 in the hairless protein, which has 1189 residues. JmjC domains have been discovered in more than 100 eukaryotic and bacterial sequences based on considerable sequence similarity, including the human hairless gene, which is mutated in people with alopecia universalis. To homology model the JmjC domain in the hairless protein, we have tried the bioinformatics method. NCBI-BLASTP, EBIClustalW, SMART, 3D-PSSM, DeepView/Swiss-PDB Viewer, PyMOL, and WhatCheck are the tools and programs utilized in this work. Using the template crystal structure of the likely antibiotic production protein from Thermus thermophilus HB8, the structure of the JmjC domain is predicted. Modeled domain structure's minimized energy value was -3394.570 KJ/mol. The simulated domain structure was validated using the WHAT IF-Proteins Model Check tool.
Downloads
References
Beaudoin, G.M., and Potter, G.B. Third: Chen, S.H., DeRenzo, C.L., Zarach, J.M., & Thompson, C.C. (2001). A new nuclear receptor corepressor is encoded by the hairless gene, which is mutated in congenital hair loss syndromes. Development and Genes., 15(20), 2687-701.
Van Steensel, M., Smith, F.J., Steijlen, P.M., Kluijt, I., Stevens, H.P., Messenger, A., Kremer, H., Dunnill, M.G., Kennedy, C., Munro, C.S., Doherty, V.R., McGrath, J.A., Covello, S.P., & Coleman, C. (1999). The hr gene was excluded by cDNA and genomic sequencing, and the gene for hypotrichosis of Marie Unna is located between D8S258 and D8S298. Journal of American Human Genetics, 65(2), 413-419.
Ahmad, W., John, P., Aslam, M., Rafiq, M.A., Amin-ud-din, M., & S. (2005). Two consanguineous Pakistani families had atrichia with papular lesions brought on by mutations in the human hairless gene. in: Archives of Dermatological Research, 297(5).
Wadhwa, G., Prakash, A., & Bhagavathi, S. (2012). An in-silico method to assess the evolutionary relationships of important proteins involved in lung cancer. Journal of Applied Pharmaceutical Science, 2(11), 084-091.
Arul, L., Balasubramanian, P., Mohan, T.M., Jayakanthan, M., Wadhwa, G., & Sundar, D. (2009). Medication design employing computer assistance for the cancer-causing H-Ras p21 mutant protein. Drug Design and Discovery Letters, 6(1), 14–20.
Wadhwa, G., Khan, A.U., & Baig, M.H. (2011). Analysis of interactions between newly discovered SHV-variants and new generation cephalosporins using molecular docking. Bioinformation, 5(8), 331-335.
S.F. Altschul, W. Gish, W. Miller, E.W. Myers, & D.J. Lipman. (1990). Simple tool for local alignment searches. Molecular Biology Journal, 215(3), 403–410.
Copley, R.R., Doerks, T., Ponting, C.P., & Bork Schultz, J. (2000). A web-based tool for researching genetically mobile domains is called SMART. Research on Nucleic Acids, 28(1), 231–234.
Gibson, T.J., Higgins, D.G., & Thompson, J.D. (1994). Through sequence weighting, position-specific gap penalties, and weight matrix selection, CLUSTAL W increases the sensitivity of progressive multiple sequence alignment. Nucleic Acids Research, 22(22), 4673-468.
Sternberg, M.J., MacCallum, R.M., & Kelley, L.A. (2000). The 3D-PSSM program's structural characteristics are used to enhance genomic annotation. Molecular Biology Journal, 299(2), 499-520.
N. Guex and M.C. Peitsch. (1997). The Swiss-PdbViewer and SWISS-MODEL: A setting for comparative protein modeling. Electrophoresis, 18(15), 2714-2723.
Verma, D.K., Gautam, B., Singh, N., Kanojia, H., Singh, S., & Wadhwa, G. (2013). Streptococcus pyogenes metabolic pathway analysis was used to estimate the structure of the drug target. Journal of Biological Science and Bioinformatics International, 1(1), 79-85.
Vriend, G. (1990). A molecular modeling and drug development program WHAT IF. Molecular Graphics Journal, 8(1), 52–56.
Downloads
Published
How to Cite
Issue
Section
ARK
License
Copyright (c) 2023 Eastman, P., Teicholz, C.

This work is licensed under a Creative Commons Attribution 4.0 International License.
Research Articles in 'Applied Science and Biotechnology Journal for Advanced Research' are Open Access articles published under the Creative Commons CC BY License Creative Commons Attribution 4.0 International License http://creativecommons.org/licenses/by/4.0/. This license allows you to share – copy and redistribute the material in any medium or format. Adapt – remix, transform, and build upon the material for any purpose, even commercially.










